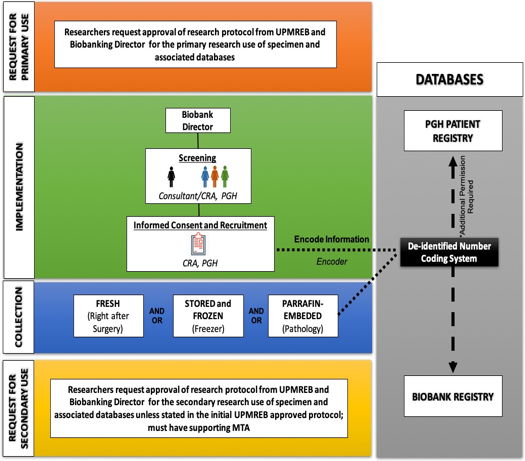
Workflow of the PGH Biobank and Associated Databases
1. Request for the Primary Research Use of Human Biological Specimens.
Researchers must request approval of the research protocol from UPMREB and Biobanking Director for the primary research use of specimen and associated databases. Most human biological specimens come from samples collected for diagnostic or therapeutic procedures, but other sources can include autopsies, volunteer donors, or materials collected and shared by other researchers. The term “biospecimen” includes sub-cellular components such as DNA or RNA, cells or tissues from any part of the human body, organs, bodily products such as hair, urine, etc., blood and blood fractions, saliva and buccal cells (Exceptions: Organisms, such as bacteria and viruses, isolated from human specimens are not human biological specimens)
2. Implementation of the Biobank.
An initial screening of patients will be done by the CRA and consultants. After confirming the diagnosis of the patient, informed consent will be offered to the patients and/or volunteers. If they consented, sample collection will proceed.
3. Collection and Storage of Biospecimens.
Depending on the type of specimen, CRAs are usually given aliquots or sections of a specimen in a collection and not the entire specimen. The various formats for collecting and storing biospecimens include: aspirates, tissue culture, frozen samples, formalin-fixed or paraffin-embedded tissues, histological slides, and extracted DNA and RNA. For the purpose of this biobank, biobank specimen will be categorized as (a) fresh samples, (b) stored/frozen, and (c) paraffin-embedded. For fresh samples, biospecimens must be obtained as fresh as possible and must be coordinated with the Primary Researcher as indicated in their approved UPREB. For frozen samples, biospecimens must be kept in the freezer immediately. For paraffin-embedded samples, biospecimens may be obtained from the pathology department. Results must be coordinated by the CRA afterward.
4. Database and Information Management
This is crucial to track specimens and any associated data. Information is usually managed by a group of specific data fields, such as, Specimen code/ID number, Specimen storage unit location, Specimen type, condition and amount, Diagnosis, Demographics, Histopathology, Patient treatment/outcome, Verification of informed consent, Donor restrictions on specimen use. This information must be stored in a secured biobank registry, different from the PGH Patient Registry. All information in the biobank registry must already be de-identified. Personal identifying data (direct identifiers) must not be stored in the biobank registry. Access and use of personal identifying data (e.g. names and addresses) from the PGH Patient Registry must be restricted and must be approved first by the UPREB.
The CRAs, encoders, and biobank chief will manage human specimen data. Researchers who obtain specimens from tissue banks and repositories often receive samples with a “limited data set.” This is to protect the identity of the subject/patient without compromising the goals of conducting meaningful research. A limited data set must have all the direct identifiers removed, such as: admission, discharge, and service dates; year of birth, and if applicable, death; age (including age 90 or over); and five-digit zip code or any other geographical subdivisions, such as state, county, city, precinct and their equivalent geocodes (except street address), treatment response and outcome data, family history information (i.e. cancer risk, gene mutation, etc.).
*Other investigators who would want access to “limited data set” should ask permission with the UPMREB and biobank director and must be legally and ethically obligated to protect data that is considered private information.
5. Request for Secondary Research Use of Human Biological Specimens
Researchers must obtain a separate approval of their research protocol from UPMREB and they must ask permission from the Biobank Chief for the use of specimen and associated databases.



